Orthology Based Substrate Prediction
This tool assigns your enzyme sequence to a pre-computed orthologous group within a chosen enzyme family, then refines that assignment by matching against only the characterized members of that group to reveal their known substrates.
In our lab’s previous work on BAHD acyltransferases, we showed that orthologous group members tend to share acceptor promiscuity accross broad compound classes (10.1111/tpj.15902). While laying cruicial groundwork, this level of activity prediction presents only an abstract idea of substrates to assay with an unknown candidate enzyme.
Recently, our lab designed a large language model workflow that extracted thousands of experimentally validated plant enzyme activities from journal articles (10.1093/bioinformatics/btae756). By leveraging the resulting curated functional data, this tool provides experimentalists with a targeted shortlist of likely substrates to test, narrowing the search space. Our goal is to extend beyond class-level predictions to improve specificity and accelerate enzyme discovery and annotation.
Method Steps:
- Input: The query sequence and target enzyme family are submitted.
- OG Mapping: An initial BLAST against the chosen enzyme family is performed to identify the query’s orthologous group.
- Focused Search: If the orthologous group contains characterized enzymes, a second BLAST is conducted using only those curated sequences.
- Annotation: For each resulting hit, key metrics — percent identity, alignment length, enzyme names, and validated substrates — are extracted.
- Output: Results are presented interactively on-page, and a TSV file with complete annotations is provided for download.
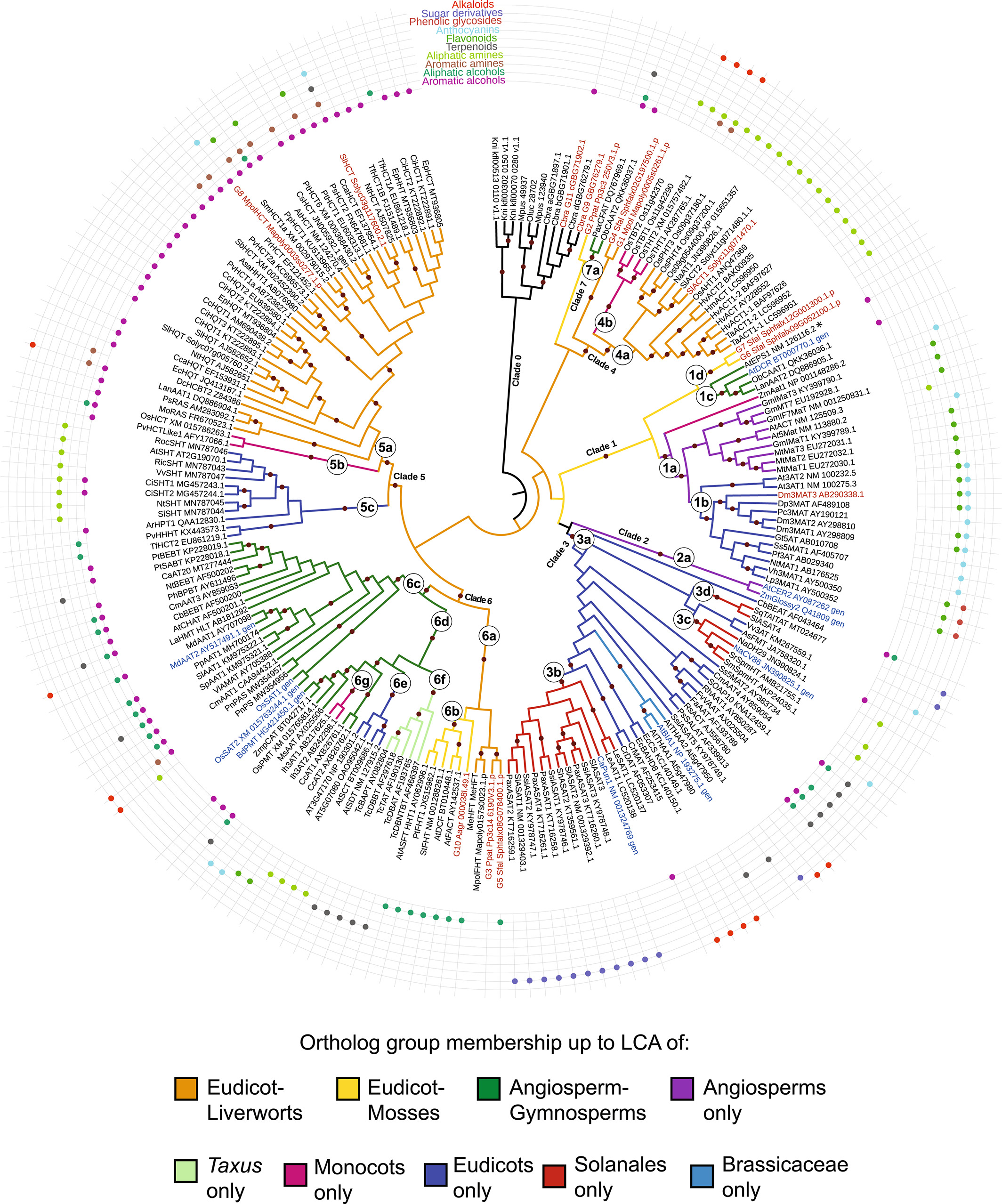
(Fig. 3 from Kruse et al. 2022)
Results: